Liquid Biopsy with Plasma-SeqSensei™
Advancements in liquid biopsy have revealed that our bodily fluids carry much more information about our health status than we previously thought. In the case of cancer, malignant cells release small traces of their DNA – called circulating tumour DNA (ctDNA) – into the bloodstream. As a result, a simple and minimally invasive blood draw can now give us insight into tumour genetic makeup. By providing information that was previously only accessible through an invasive tissue biopsy, such blood draws help closely monitor the effectiveness of ongoing cancer treatment. But not all liquid biopsy tests are created equal. At Sysmex, we strive to deliver sensitive, truly quantitative and transferable liquid biopsy technology with Plasma-SeqSensei™ kits.

Even low levels of ctDNA could have given an indication of the coming progression in the follow-up.

This Plasma-SeqSensei™ technology can offer a better chance to obtain a targeted therapy for patients with MAF below 1%
High sensitivity matters
Many cancer patients only have a small amount of cancer DNA in their blood, making it hard to detect. Studies using liquid biopsy kits from Sysmex have shown that 20–40% of patients in metastatic breast cancer, colorectal cancer, non-small cell lung cancer or pancreatic cancer have mutant allele fractions (MAFs) between 0.06-0.5%1-9.
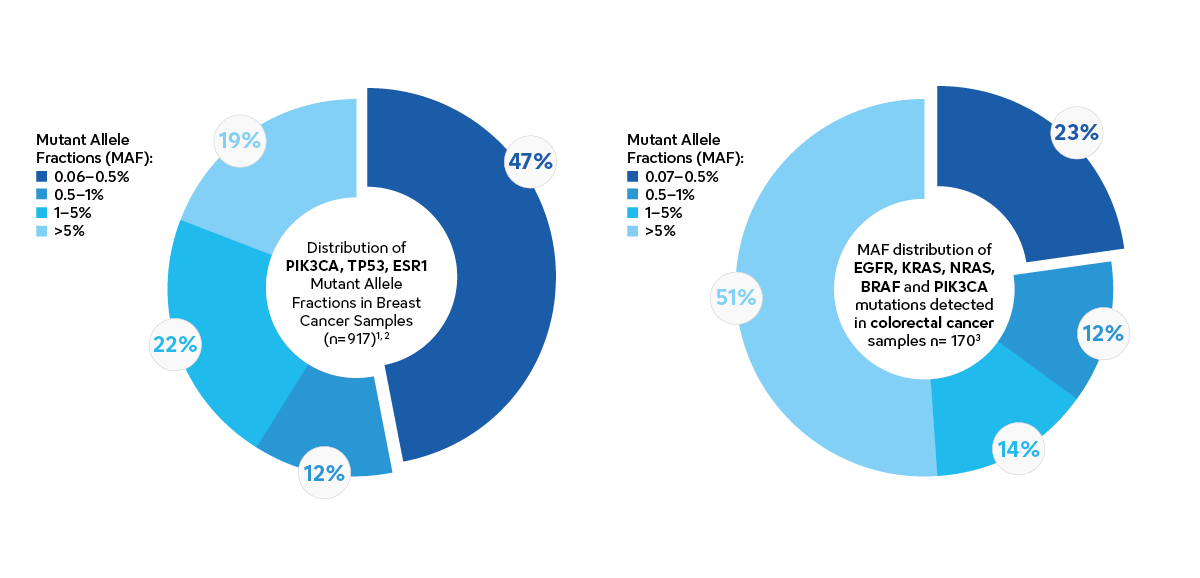
To accurately find these cancer DNA molecules, a sensitive liquid biopsy analysis is needed. However, most NGS-based liquid biopsy solutions have a sensitivity of 0.5%-1% mutant allele fractions.
Sysmex’s solution
Plasma-SeqSensei™ offers highly sensitive NGS-based liquid biopsy kits, detecting mutant allele fractions (MAFs) down to 0.06–0.07%.A
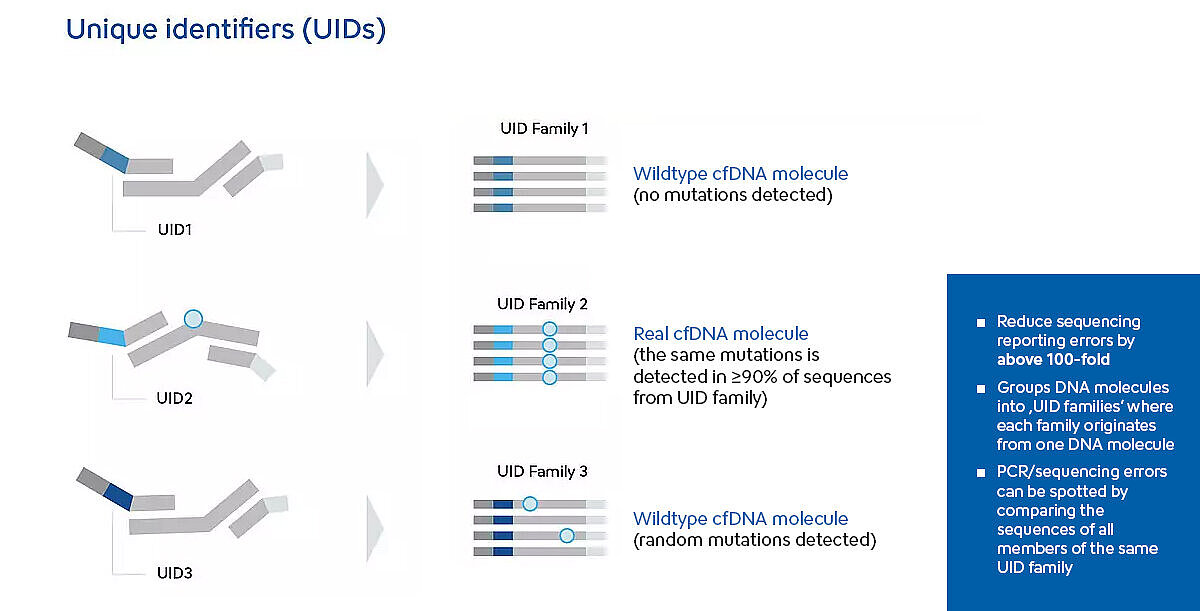
To enhance sensitivity and minimise sequencing-associated errors, the Plasma-SeqSensei™ library preparation workflow incorporates deep sequencing and SafeSEQ technology7. This technology assigns a unique identifier (UID) to each DNA molecule during the target selection step. UIDs enable precise mutation tracking and make it possible to distinguish mutations from potential polymerase or sequencing errors.

There is much more diagnostic power in the absolute quantification.
Beyond MAF – absolute mutant molecules quantification
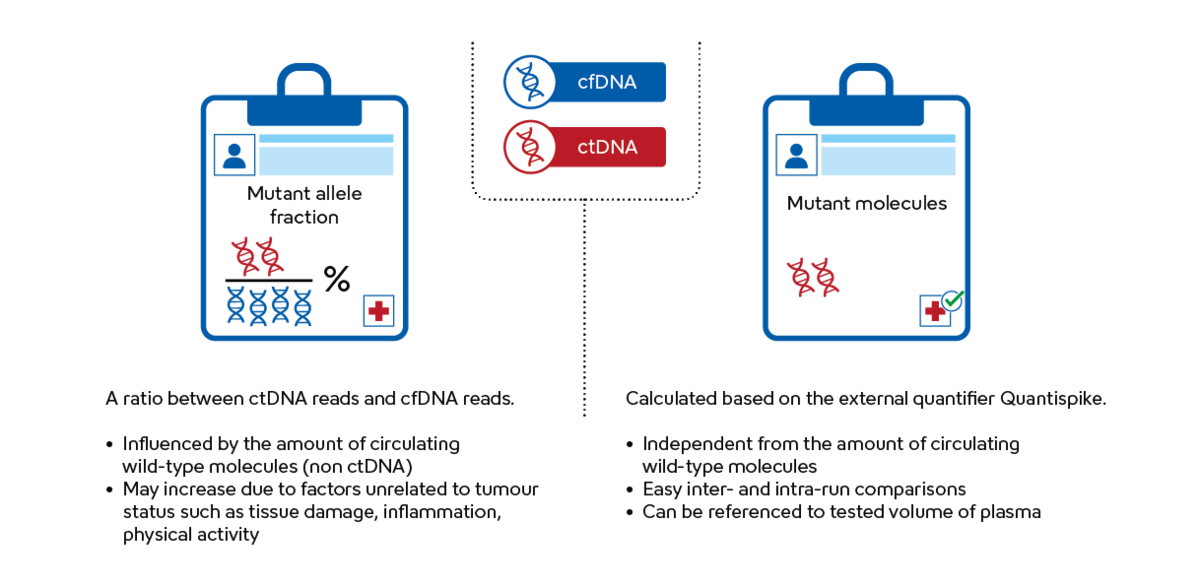
Liquid biopsy results are commonly reported as MAF values, measured as the ratio between the sequences containing mutations and all the sequences detected in the sample (wild-type and sequences containing mutations). However, the quantity of wild-type sequences can exhibit significant variability due to various factors unrelated to the tumour's status. These factors may include tissue damage, inflammation and even physical activity. This variability can introduce a level of bias into conventional NGS-based liquid biopsy measurements, impacting the accuracy of quantitative measurements, which is important for longitudinal monitoring of the patients.
Sysmex’s solution
Plasma-SeqSensei™ technology utilises Quantispike, an internal quantifier that allows quantification of tumour-specific sequences independently from the number of wild-type sequences. Absolute quantification reported as a number of mutant molecules detected in the tested volume of plasma facilitates a meaningful comparison of changes in mutation levels over time.B
Bringing NGS-based liquid biopsy in-house offers numerous advantages
Time-efficiency: reduced turnaround times enable faster access to crucial results.
Cost savings: minimise outsourcing costs while maintaining control over patient samples.
Data security: keeping sensitive data within your organisation.
Quick decision-making: rapid access to data empowers timely and well-informed diagnostic decisions.
Despite these advantages, the complexity of some NGS workflows might be an obstacle for widespread implementation into diagnostic laboratories.
Sysmex’s solution
Quick and accessible
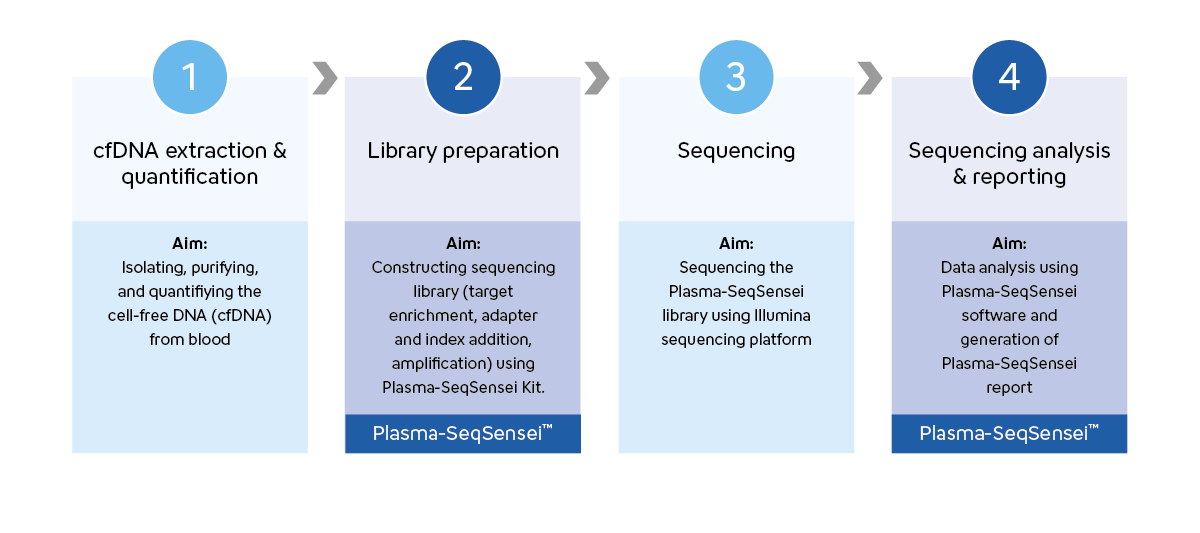
Plasma-SeqSensei™ workflow is designed to optimise laboratory processes. It offers a simple amplicon-based approach with library preparation completed in one day, followed by overnight sequencing on the Illumina platform, and comprehensive analysis and reporting using Plasma-SeqSensei™ Software on the subsequent day.
User-friendly, two-step PCR process
Our method employs a two-step PCR process that was developed to be straightforward and easy to follow, making liquid biopsy accessible to a wider range of laboratories that conduct NGS tests.
Detailed training resources
Our website provides training videos that offer complete guidance throughout the workflow. Plasma-SeqSensei™ basic online training | Caresphere Academy DACH (caresphere-academy.com)
Sysmex support for workflow implementation
We understand that implementing a new workflow can be difficult. That's why Sysmex is here to support you in bringing this technology into your laboratory. Our team is ready to assist you in the initial implementation, ensuring a smooth transition and successful integration of our NGS workflow into your daily laboratory routine.
Robust software with an intuitive interface
Tackling the complexity of NGS analysis, our Plasma-SeqSensei™ software provides a robust, IVD-certified solution with an intuitive interface, ensuring seamless data processing. Our unique software simplifies and automates data analysis while delivering precise quantification of cfDNA sample inputs.
It is designed to run effortlessly on standard office PCs, eliminating the need for specialised workstations or cloud infrastructure, making it possible to keep sensitive patient data in-house. It can even be run in an offline work environment for more data protection and security.
The software's interface is divided into three intuitive modules, simplifying the analytical process:
- Run planning: helps to streamline analysis with ease.
- Data analysis: go from FASTQ input files to results in just a few clicks.
- Reporting: enables the gathering of actionable insights from the data.
Comprehensive reporting for clinical researchers
The automatically generated, concise report includes only relevant information, including:
- Mutation status clearly indicates the presence or absence of mutations including quantification results as both mutant molecules and mutant allele fractions.
- Sample metrics provide insights into the quality and characteristics of the sample.
- Quality metrics ensure the reliability of the reported data.
Plasma-SeqSensei™ software handles the complexity of data analysis for you, allowing you to focus on insights rather than intricacies.
Disclaimers
[A] 0.06% - 0.07% MAF on a background of 10,000 GE represent the assays cutoffs of 6 resp. 7 mutant molecules (MM). Please refer to Plasma-SeqSensei™ Kit IFUs accessible on www.mysysmex.com for information on performance characteristics.
[B] The quantification of mutant molecule counts can only be accurately determined above the lower Limit of Quantitation (LOQ) Please refer to the Plasma-SeqSensei™ Kit IFUs available on www.mysysmex.com for information on performance characteristics.
CE-IVD solutions
References
[1] Martens G et al. (2025): J Mol Diagn 27(1): 25-35.
[2] FLIPPER trial (unknown): in preparation
[3] Lago et al. (2025): X Liquid Biopsy Symposium
[4] Barthelemy D. et al. (2023): Cancers 15.5: 1574.
[5] Schmiegel et al. (2017): Mol Oncol. 11(2): 208 – 219;
[6] Saunders et al. (2016): Annals of Oncology 27 (6): 149 – 206;
[7] Toledano-Fonseca M. et al (2020): Cancers 12.7: 1754.
[8] Vidal et al. (2017): J Clin Oncol 35 (suppl 4S; Abstract 607);
[9] Oxnard et al. (2016): J Clin Oncol 34(28): 3375-3382