Interpret NGS Analysis Software
Effortless translation of NGS data into meaningful results
- Unlimited-use complimentary software
- Highly configurable
- Locally installed or on the cloud
The Interpret NGS Analysis Software delivers:
- Detection of wide range of aberrations ranging from low frequency SNVs and indels to large structural deletions including CNVs and translocations
- Extensive customisation options for variant and batch reports and database links to meet the laboratory’s exact needs
- Comprehensive range of filtering options to meet your analytical criteria
- Security and control to log and track user activity and standardise analysis protocols through multiple access permission levels
- Powerful and complementary with SureSeq™ and CytoSure® NGS panels to detect all aberrations
Interpret is a powerful and easy-to-use next-generation sequencing (NGS) analysis solution, facilitating analysis and visualisation of a wide range of variants and structural aberrations. Coupled with a comprehensive and powerful filtering framework, the software delivers accurate calling of single nucleotide variations (SNVs) and indels, as well as structural aberrations, including internal and partial tandem duplications (ITDs, PTDs), copy-number variations (CNVs), loss-of-heterozygosity (LOH) and translocations. Interpret is designed to work seamlessly with all CytoSure® and SureSeq™ NGS panels and offers flexible accessibility for data analysis; whether through a stand-alone computer, laboratory server or another web-enabled device.
Complementing the expert panel design and hybridisation-based approach of NGS panels to deliver unparalleled coverage uniformity, Interpret is integral in facilitating the detection of low-frequency variants consistently and with confidence. With a wide range of customisation options and links to various mutation databases, Interpret provides effortless translation of NGS data into meaningful results. Additional outputs from the pipeline include a range of publicly available data resources for annotation of variants detected, including ClinVar, COSMIC, dbSNP, Ensembl, gnomAD, PolyPhen, SIFT. Additional licence-based sources, such as HGMD, can be incorporated on provision of suitable credentials.
Using Interpret, samples are rapidly processed through customisable protocols and optimised settings to generate the variant lists. Your laboratory has the option to use the protocols provided with the software or develop your own through the intuitive user interface. Personal modifications are available for: hardware settings, quality metrics, variant calling parameters and, variant filtering parameters.
Following analysis, results can be viewed in the user-friendly variant browser showing a tabular display of the calls and an IGV window, which can be maximised in a separate window for greater visibility. Report generation is implemented through a templating system. Interpret report templates are highly customisable and can be designed to individual requirements.
SNVs
Following analysis, all variants are displayed in a table, below which is an Integrative Genomics Viewer (IGV)1 window allowing a more detailed review of the data and additional verification.
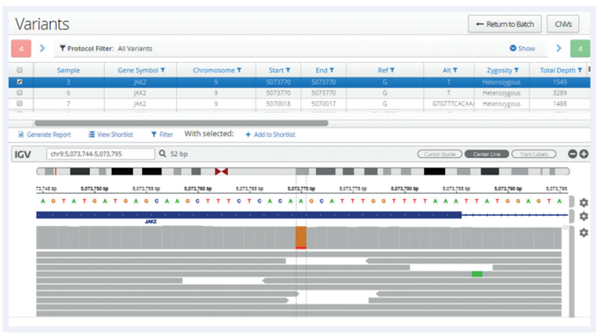
Figure 1: In this example a low-frequency JAK2 V617F SNV has been selected and the user is able to view the aligned reads generated by the pipeline.
ITDs
Detection of FLT3-ITDs of various sizes, including regions containing multiple ITDs. Note how Interpret can confidently identify even ITDs much longer than the sequencing read length of 150 bp.

Figure 2: ITD sizes are (A) 174 bp, (B) 225 bp, (C) 195 bp with an additional 6 bp, (D) 120 bp and (E) 168 bp with an additional 69 bp.
PTDs
In conjunction with SureSeq™’s expert panel design, Interpret offers robust detection of all sizes of PTDs in KMT2A.
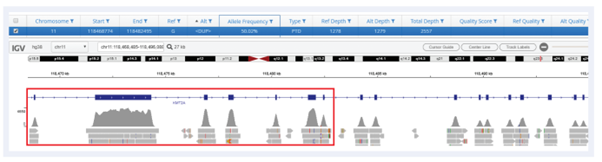
Figure 3: Detection of a KMT2A-PTD spanning exons 2-8.
Translocation
Interpret agnostically detects split-reads across the genome, enabling detection of both known and unknown translocation partners.

Figure 4: BCR-ABL translocation. Split-reads covering both BCR (left panel) and ABL1 (right panel) are detected, indicative of the BCR-ABL gene fusion.
CNVs
Interpret enables CNV detection ranging from loss of a single exon to full chromosomal arms and trisomies.
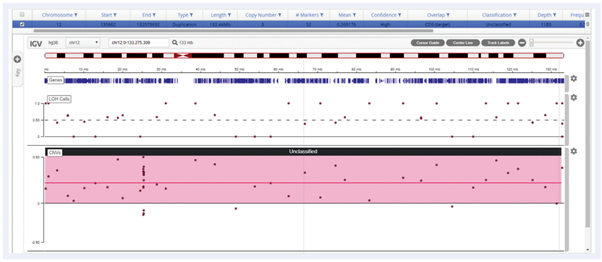
Figure 5: Detection of trisomy 12 using the SureSeq™ CLL + CNV V3 Panel, showing a reliable gain call across the whole chromosome.
LOH
CytoSure® Constitutional NGS panel analysed using Interpret.
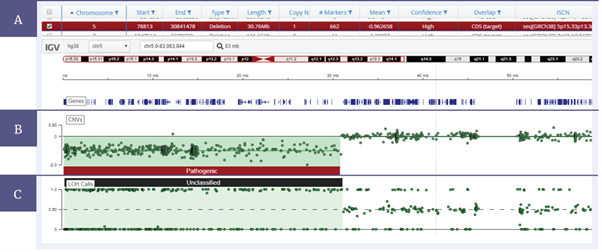
Figure 6: 30 Mb deletion on chromosome 5 and LOH across the deletion. (A) Details of the samples and mutations are displayed in a table format. (B) The CNV data is displayed in a log2 ratio plot and (C) a b-allele plot shows the LOH present within the sample.
| Basic Variant Attributes | Variant Call Attributes | Consequence/ Severity Predictions | Population Frequencies | Classification/ Pathogenicity | Genes, Exons and Proteins | Region/ Variant Lists |
Chromosome | Total Depth | Most Severe Consequence | rsID | ClinVar Significance | Gene ID | Region Lists |
Start | Ref / Alt Depth | Impact | Minor Allele / Freq | Gene Symbol | Variant Lists | |
End | Allele Frequency | Terms | American Minor Allele / Freq | Transcript ID | ||
Genome Build | Quality Score | PolyPhen Prediction | European Minor Allele / Freq | Protein ID | ||
Ref | Ref / Alt Quality | PolyPhen Score | African Minor Allele / Freq | Exon ID | ||
Alt | Log Ratio | SIFT Prediction | South Asian Minor Allele / Freq | Exon Number | ||
| Genotype | Ref Reads (+) / (-) | SIFT Score | East Asian Minor Allele / Freq | |||
Alt Reads (+) / (-) | ||||||
Ref Strand Bias | ||||||
| Reads Placed Left / Right |
Table 1: Overview of the wide range of filtering options available in Interpret.
References:
1. Helga Thorvaldsdóttir, James T. Robinson and Jill P. Mesirov. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14(2), 178-192 (2013).
Disclaimer:
SureSeq™ and CytoSure®: For Research Use Only, not for use in diagnostic procedures. Product availability may vary from country to country. Contact your local representatives for availability.
Manufacturer and Trademarks: SureSeq™ and CytoSure® (Oxford Gene Technology IP Limited)
| Requirement | Minimum | Recommended |
| Memory (RAM) | 16 GB | 32 GB (64 GB for MRD) |
| CPU cores | 8+ logical cores at 2+ GHz | 16 CPUs (36 CPUs for MRD) |
| Storage (disk space) | 500 GB | 2 TB |
| Operating system | Windows 10 Pro or newer (+ Virtual Machine) Any Unix supporting Docker CE (no VM) | |
| No requirements for graphic cards | ||
| Virtualisation (VT-x) to be enabled in the BIOS |
Minimum and recommended hardware requirements for running Interpret both locally and on Cloud.
Sysmex Europe SE
Deelböge 19 D
22297 Hamburg
Germany
+49 (40) 527 26 0
+49 (40) 527 26 100
Product documents
Regulatory documents
Regulatory documents, such as Instructions for Use, can be accessed with a valid My Sysmex login:
Go to My Sysmex